Sequence Tolerance Server Documentation
Overview
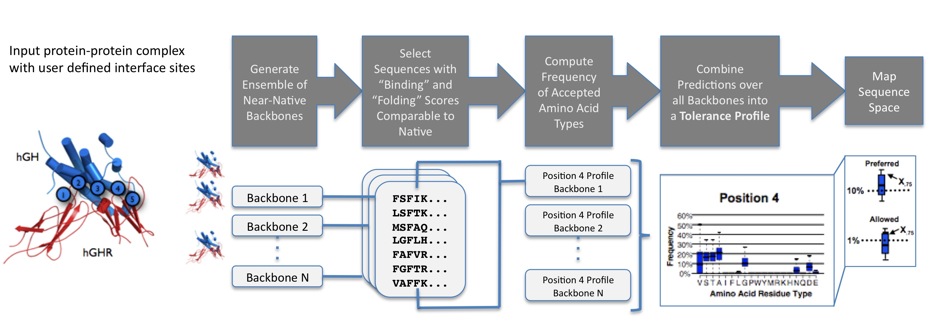
The Sequence Tolerance application predicts the relative frequencies with which amino acid residues are predicted to be "tolerated" without significantly compromising the stability of a given protein-protein interface and the respective interacting partner proteins. The process is as follows:- The user defines a small number of interface sites (typically 4-6) at which mutations to all 20 naturally occurring amino acids (except cysteine) will be allowed in the simulations.
- In a first step, Rosetta calculates an overall interface binding score and a fold stability score for each protein chain in the given protein-protein complex.
- Next, Rosetta models and scores non-native sequences at the interface sites selected for design by the user.
- Non-native sequences at the protein-protein interface are selected and propagated by a genetic algorithm; for each sequence, the rotameric conformations of each individual sequence at the interface are optimized by a Monte Carlo simulated annealing protocol and the resulting structural models are scored using the all-atom Rosetta scoring function.
- Throughout the simulation, non-native sequences with folding and binding scores near or significantly better than the starting sequence are saved and considered to be "tolerated".
- At the end of a simulation, the frequency with which each amino acid type appeared in the list of tolerated sequences is calculated and used to generate a position-specific "tolerance profile" for each interface site.
 Sequence plasticity predictions more closely mimic experimentally determined interface tolerances when made over an ensemble of backrub generated backbones.
Sequence plasticity predictions more closely mimic experimentally determined interface tolerances when made over an ensemble of backrub generated backbones.
- When using an ensemble of structures, sequence plasticity predictions are generated independently for each member in the ensemble by using the process outlined above.
- For each interface position, individual position-specific "tolerance profiles" for each backbone in the ensemble are combined and represented in box-plot format.
- Some amino acid substitutions within a protein-protein interface will show very low predicted frequencies for all members among an ensemble and can be considered forbidden, or not allowed.
- The frequency at which other mutations are predicted may vary depending on which backbone within the ensemble is used. This backbone variation may indicate mutations predicted to be tolerated at a protein-protein interface only when certain small backbone adjustments are considered.
Sequence Tolerance basic application
Tips:
- The application works best at up to six designed residues. The length of processing is tailored to sample sufficiently for this number of designed positions. Using more than six designed positions is currently discouraged.
Inputs:
You need to input:
- Starting pdb file
- At least one designed position
Interpreting Results
- Todo:
Citations
We would be grateful if you cite our work in publications that make use of the server:
- Smith CA, Kortemme T (2010).Structure-Based Prediction of the Peptide Sequence Space Recognized by Natural and Synthetic PDZ Domains. Journal of Molecular Biology, Volume 402, Issue 2, 17th September 2010, Pages 460-474.
- Smith CA, Kortemme T (2011).Predicting the Tolerated Sequences for Proteins and Protein Interfaces Using Rosetta Backrub Flexible Backbone Design. PLoS ONE 6(7): e20451. doi:10.1371/journal.pone.0020451
More questions?
We welcome scientific and technical comments on our server. For support please contact us at Rosetta Forums with any comments, questions or concerns.