Supercharge Server Documentation
Intro
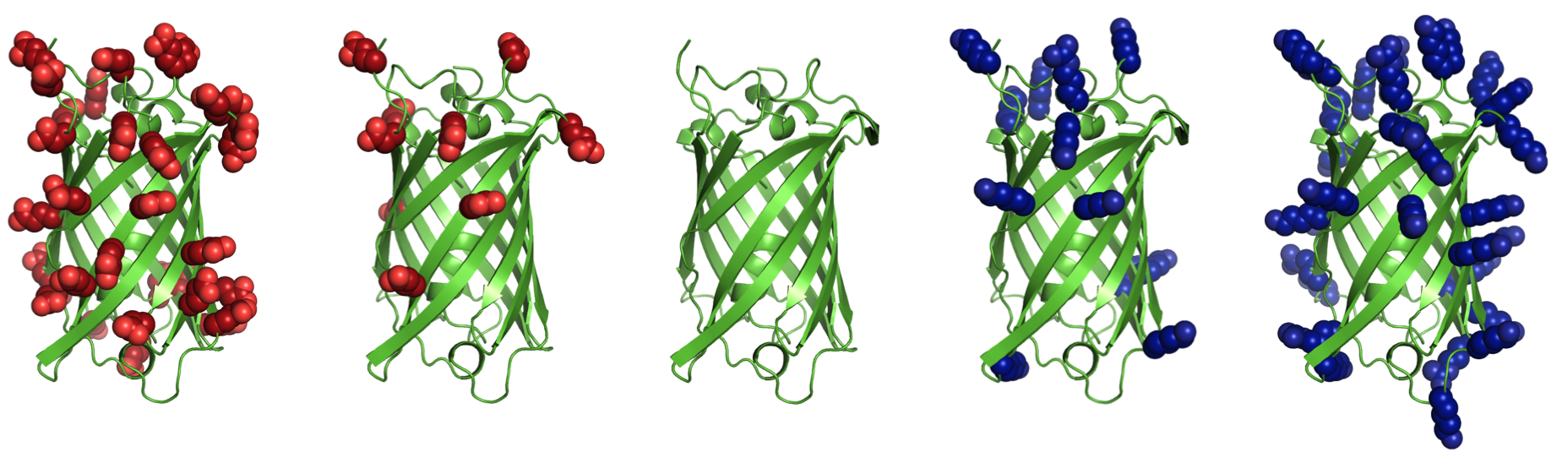
Supercharging refers to increasing the net charge of a protein by mutating surface residues. Increasing net charge can improve reversibility of unfolding by preventing aggregation of partially unfolded states. Aggregation is a common obstacle for use of proteins in biotechnology and medicine. Net charge, rather than number of charged residues, is often an indicator of aggregation propensity. Additionally, highly cationic proteins and peptides are capable of nonviral cell entry, and highly anionic proteins are filtered by kidneys more slowly than neutral or cationic proteins.Supercharge application
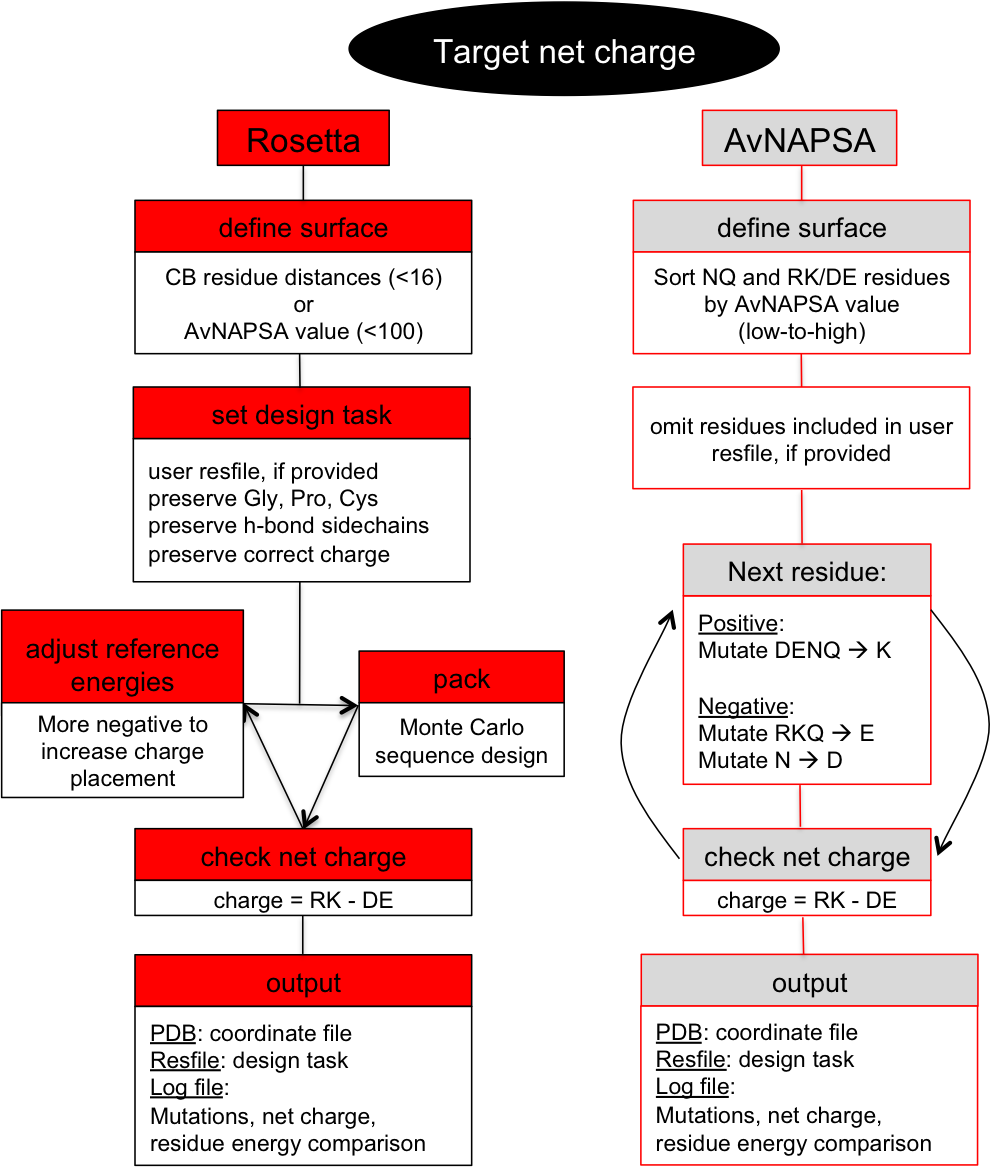
This is a surface redesign protocol. Optimal positions for incorporation of charged side chains should be determined, as numerous mutations and accumulation of like-charges can also destabilize the native state. A previously demonstrated approach (AvNAPSA) deterministically mutates flexible polar residues (amino acids DERKNQ) with the fewest average neighboring atoms per side chain atom. Our approach uses Rosetta-based energy calculations to choose the surface mutations. Both automated approaches for supercharging are implemented in this protocol.The AvNAPSA approach varies net charge by adjusting the surface cutoff. The Rosetta approach varies net charge by adjusting reference energies of the positive or negatively charged residues.
Supercharging does not predict the optimal net charge for a given application, it is intended to generate a series of sequences with different mutations and net charges for experimental testing.
AvNAPSA-mode
AvNAPSA supercharge philosophy: mutate the most exposed polar residues to minimize structural change or destabilization. Only DE-RK-NQ residues can be mutated. This final sequence is deterministic.Drawbacks: mutating surface polar residues can eliminate hydrogen bonds. Helix capping, edge-strand interaction, and loop stabilization all result from surface hydrogen bonds. Furthermore, this automated protocol mutates N to D and Q to E, but N and Q sometimes act simultaneously as a donor and acceptor for hydrogen bonds.
Reference: Lawrence MS, Phillips KJ, Liu DR. Supercharging proteins can impart unusual resilience. J Am Chem Soc. 2007 Aug 22;129(33):10110-2.
- AvNAPSA-mode with a target net charge
- Define surface. sort NQ and RK/DE residues by AvNAPSA value (low to high).
- Next residue in sorted list: Positive: mutate DENQ-->K, Negative: mutate RKQ-->E and N-->D.
- If net charge = target net charge, output pdb.
- AvNAPSA-mode with a surface cutoff
- Define surface by AvNAPSA value (less than 100 for moderate charging, less than 150 for heavy charging.
- For each NQ and DE/RK residue in the surface: Positive: mutate DENQ to K, Negative: mutate RKQ to E and N to D.
- Output pdb.
Rosetta-mode
Rosetta supercharge philosophy (Rsc): mutate residue positions that preserve and/or add favorable surface interactions. Hydrophobic and small polar surface residues can also be mutated.Rosetta drawbacks: mutating less-exposed positions can lead to better computed energies, but mistakes at these positions can be destabilizing. AvNAPSA favors charge swaps, so Rosetta requires more mutations to accomplish the same net charge.
Reference: Miklos AE, Kluwe C, Der BS, Pai S, Sircar A, Hughes RA, Berrondo M, Xu J, Codrea V, Buckley PE, Calm AM, Welsh HS, Warner CR, Zacharko MA, Carney JP, Gray JJ, Georgiou G, Kuhlman B, Ellington AD. Structure-based design of supercharged, highly thermoresistant antibodies. Chem Biol. 2012 Apr 20;19(4):449-55.
- Rosetta-mode with a target net charge
- Define surface. Neighbor by distance calculator (CB dist.), less than 16 neighbors default or Define surface by AvNAPSA value (less than 100 default)
- Set design task
- read user resfile, if provided
- use Arg/Lys/Asp/Glu
- dont_mutate gly, pro, cys
- dont_mutate h-bonded sidechains
- dont_mutate correct charge residues
- Set reference energies for RK/DE, starting at user input values
- pack rotamers mover
- check net charge, increment/decrement reference energies (back to step 3.)
- Once a pack rotamers run results in the correct net charge, output pdb
- Rosetta-mode with surface cutoff and input reference energies for the desired charged residue types (Arg/Lys for positive-supercharging, Asp/Glu for negative-supercharging)
- Define surface. Neighbor by distance calculator (CB dist.), less than 16 neighbors default or Define surface by AvNAPSA value (less than 100 default)
- Set design task
- read user resfile, if provided
- use Arg/Lys/Asp/Glu
- dont_mutate gly, pro, cys
- dont_mutate h-bonded sidechains
- dont_mutate correct charge residues
- Set reference energies for RK/DE, using the user input values pack rotamers mover
- Output pdb
Supercharge options
The only required input file is the PDB to be redesigned. If homology models, NMR ensembles, or relaxed crystal structures are the starting structure, -l can be used.
Optionally, the user can use a resfile to specify residue positions to NOT mutate (NATAA or NATRO). This would be useful to preserve a known binding surface, for example. The default for the input resfile should be ALLAA and the supercharge protocol restricts the allowed residues at designable positions. For example, Gly, Pro, Cys residues and hbonded sidechains are not allowed to mutate by default.
- -------------------Options, AvNAPSA and Rosetta modes:-------------------
- target_net_charge SIGNED_INT def(0) //AvNAPSA: Residue positions will be mutated one at a time from most exposed to least exposed until the target net charge is achieved. Rosetta: reference energies increment until desired net charge results from Monte Carlo sequence design
- surface_definition_atom_neighbor_cutoff UNSIGNED_INT def(100) // this is how AvNAPSA defines surface, can be used in either approach
- compare_energies BOOL def(false) //prints a full residue-by-residue energy analysis in the log file
- only_compare_mutated_residues BOOL def(false) //only includes mutated residues in the energy analysis
- resfile FILE // this is how you can specify which residues to not mutate. Default setting must be ALLAA, and residue-by-residue settings should be NATAA, as shown below: ALLAA
- -------------------Options, AvNAPSA mode:-------------------
- AvNAPSA_positive BOOL def(false) //run positive-charge AvNAPSA
- AvNAPSA_negative BOOL def(false) //run negative-charge AvNAPSA
- surface_definition_atom_neighbor_cutoff UNSIGNED_INT def(100) // this is how AvNAPSA defines surface, can be used in either AvNAPSA or Rosetta modes. less than 100 is good for light supercharging, 150 is good for heavy supercharging. Rosetta typically defines surface according to number of residue neighbors (16 default). Thte atom-based and residue-based surface definitions correlate with an R-squared of 0.85, so they are moderately similar.
- -------------------Options, Rosetta mode:-------------------
- surface_definition_by_atom BOOL def(false) //Tell Rosetta-mode to use AvNAPSA values to define the surface. The default, however, is to use number of residue neighbors cutoff.
- surface_definition_residue_neighbor_cutoff UNSIGNED_INT def(16) //residues with 16 neighboring residues within 10 Å are considered part of the surface
- include_arg BOOL def(false) //use arginine in Rosetta supercharge
- include_lys BOOL def(false) //use lysine in Rosetta supercharge
- include_asp BOOL def(false) //use aspartate in Rosetta supercharge
- include_glu BOOL def(false) //use glutamate in Rosetta supercharge //the reference energies of the charged residue types will govern the net charge of Rosetta designs. Rosetta can choose between the allowed charged residue types and the native residue. More negative reference energies will result in more charge mutations. The user can increment reference energy values to vary the resulting net charge. For positive-charging, refweight_asp and refweight_glu values will be ignored. For negative-charging, refweight_lys and refweight_arg values will be ignored.
- refweight_arg FLOAT def(-0.98)
- refweight_lys FLOAT def(-0.65)
- refweight_asp FLOAT def(-0.67)
- refweight_glu FLOAT def(-0.81)
- dont_mutate_glyprocys BOOL def(true) //glycine, proline, and cysteine often serve special structural roles in proteins, these are not mutated by default.
- dont_mutate_correct_charge BOOL def(true) //i.e., Don’t mutate arginine to lysine. Don't mutate aspartate to glutamate.
- dont_mutate_hbonded_sidechains BOOL def(true) //don’t mutate residues with sidechains forming a hydrogen bond
- pre_packminpack BOOL def(false) //Packrotamers is always done as the first step. This option will go one step further and run packrotamers, sidechain+backbone minimization, packrotamers on the input structure before performing the supercharge design step.
- nstruct UNSIGNED_INT def(1) //Monte Carlo sequence design of a protein surface is often convergent but it is still stochastic, multiple design runs can be performed if desired.
start
20 A NATAA
24 A NATAA
26 A NATAA
if the default were NATRO, for example, no design would occur!
Tips
For homology model ensembles or NMR ensembles, we recommend supercharging all the input structures and choosing consensus mutations. For supercharging a single crystal structure, or ensembles, we recommend generating designs with a spectrum of net charges. Since repacking the surface converges on similar sequences, we do not recommend using nstruct more than 10 (actually, nstruct is not used for AvNAPSA-mode because the sequence is deterministic). When positive-supercharging, you can bias the choice of Arg vs. Lys by giving different reference energies for the two residues. Likewise for negative-supercharging. The protocol dumps output PDBs with customized self-documenting names.Outputs
- Output 1: PDB
- Output 2: resfile. This is an output resfile (rather than input) so the user can see what the design task looked like.
- Output 3: log. The output includes net charge, number of mutations, what the mutations are, pymol selection for convenient viewing of mutated residues, and a residue-by-residue energy comparison of the designed vs. starting structures.